Bruker Daltonics, September 2020
Keeping employees safe while supporting customers during the Covid-19 pandemic
All Bruker facilities are keeping employees safe, including the Billerica, Massachusetts demo facility. Take a look at how we are working safely to provide support and demonstrations while maintaining all safety requirements during COVID-19.
| Watch Video |
|
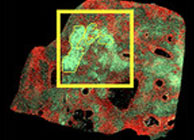
MS Imaging-Guided Microproteomics for Spatial Omics on a single instrument
Use SpatialOMx® to subtype tumors by molecular phenotype then select and target subtypes for deeper 4D-omics analysis. Prof. Ron Heeren’s team at the M4I has published a new study using SpatialOMx® to capture deep proteomic diversity of different lipid subtypes of tumors.
| Link to Full Article |
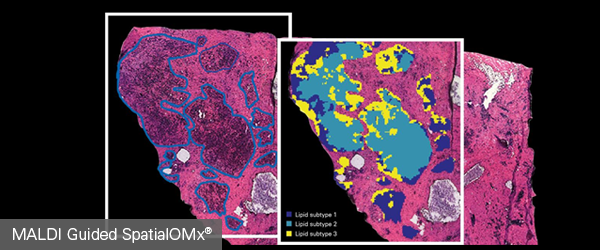
SpatialOMx® combines molecular imaging and 4D-Omics to probe tissue substructures
In order to understand how individual cells act and interact in the context of their heterogeneous tissue environment, it is imperative to examine their molecular profiles in situ. However, traditional analysis tools are limited, either disregarding the spatial component as with homogenization followed by LC-MS, or monitoring a very limited range of proteins as with immuno-based imaging. MALDI Guided SpatialOMx® allows subtyping of tissue and even further subpopulation of cells can be subsequently targeted for 4D-OMICS analysis to extract deepest molecular insight with spatial context.
| Learn More |
|
Uncover proteomic profiles in tumor subpopulations
MALDI Guided SpatialOMx® enables you to focus your X-Omics analyses onto cells expressing a specific molecular phenotypes that may not even differentiate histologically. Bruker’s timsTOF fleX can segment tissue into cellular subpopulations using molecular expression, that allows you to locate certain molecular phenotypes for selective targeting by deep X-Omic analysis.
| Link to Full Article |
|
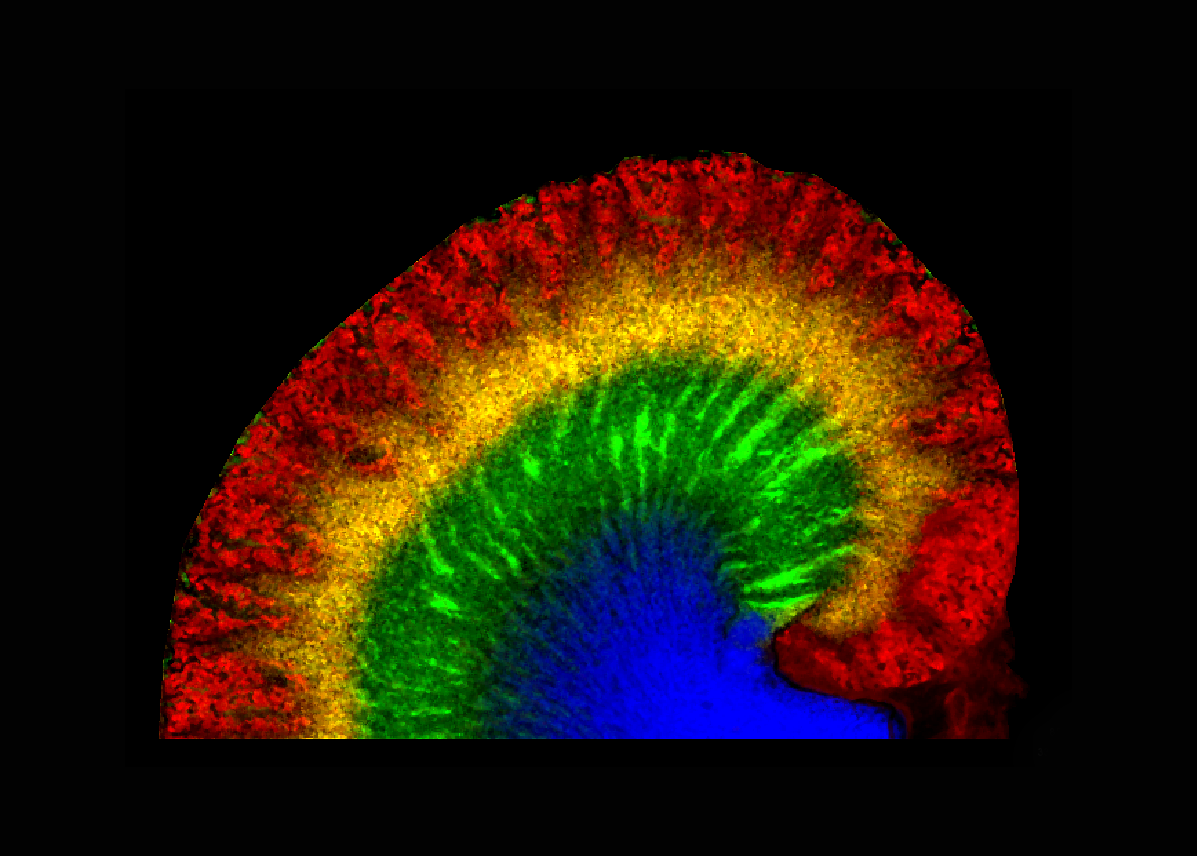
Capture greater molecular insight into the complex tissue environment
In this presentation Prof. Ron Heeren, Division Head of Imaging Mass Spectrometry at University of Maastricht, describes how SpatialOMx® provides greater specificity and sensitivity for understanding cellular expression in spatial context.
| Watch Presentation |
|
Guide for writing successful proposals for SpatialOMx® and timsTOF flex
MALDI Guided SpatialOMx® represents a quantum advance in how tissue is analyzed. It combines a label-free imaging technique that delivers sensitive distribution mapping for both targeted and untargeted compounds, ranging from metabolites to lipids to proteins with regionally targeted 4D-Omics analysis.
| Learn More |
|
John R. Yates, III and Robin Park from the Scripps Research Institute
Mass spectrometry has always had a powerful synergy with computers. Computers have pushed mass spectrometry forward at key junctures in its history from data collection to instrument operation to data analysis. Proteomics was enabled by both tandem mass spectrometry and informatics to rapidly assign amino acid sequences to spectra. As instrumentation has become more powerful informatic capabilities have grown to keep pace with increases in data production and data types. Sophisticated workflows are used to process proteomic experiments that encompass search, quantitation, and statistical processing of data.
As new features are added to mass spectrometers like ion mobility this provides additional capability for collecting data and information for interpreting peptides and peptide features. IP2 is a proteomic platform that creates a workflow combining GPU powered search, flexible quantitation, and statistical analysis of data.
| Learn More |
|
High-througput 4D-Proteomics™
Read about the benefits of dia-PASEF technology on the timsTOF Pro platform coupled to an Evosep One for high-throughput in-depth proteome analysis for short gradients. The timsTOF Pro coupled with the Evosep is best in-class running up to 300 samples per day.
| Read More |
|
Proteomic Interrogation of Primary Insulin-secreting Pancreatic β-cells
Dr. Jarrod Marto’s lab from the Dana-Farber Cancer Institute recently published a paper in Nature Metabolism on a multi-omic characterization of Primary Insulin-secreting Pancreatic β-cells. A summary application note of the proteomic analysis performed with the timsTOF Pro can be found here. The proteome depth given limited sample amounts is amazing.
| Read More |
|
CCS values to enable 4-dimensional annotation of metabolic features
Reliable annotation of features is essential for metabolomics research. This new application note generated in collaboration with the team from Metabolomics pioneer Prof. Jeremy Nicholson highlights how reproducible CCS values enable high confidence metabolite assignment – even across continents.
| Read More |
|
4 Reasons to switch to 4D-Proteomics™ on the timsTOF Platform
Four reasons to switch to 4D-Proteomics™ on the timsTOF platform. To provide concise arguments for replacement of older 3D mass spectrometers with the 4D capable, timsTOF platform and its unique TIMS (Trapped Ion Mobility Spectrometry) capability which adds the CCS dimension to retention time, precursor mass, and MS/MS.
| Learn More |
|
|
Virtual Events
ASMS 2020
HUPO CONNECT
PHARMA Webinar Tour
|
Latest media from Bruker
Follow Us